(Co)evolution at the interface of species interactions
Hague lab
RESEARCH

A cellular-genetic basis for endosymbiont prevalence
Endosymbioses are intimate host-microbe relationships where bacteria reside within the cells of their host. Endosymbionts alter basic aspects of host biology, evolution, and ecology. We study the spread and evolution Wolbachia, the most common endosymbionts in nature. Wolbachia infect about half of all insects species and biocontrol programs employ virus-blocking Wolbachia to prevent mosquitoes from transmitting deadly diseases like dengue. Our research goal is to understand how cellular host-microbe interactions determine global Wolbachia prevalence and coevolution with hosts.
Transmission rates help explain Wolbachia prevalence
The prevalence of Wolbachia and other endosymbionts varies widely across environments and insect species. Wolbachia are vertically transmitted from mother to offspring; however, this process can breakdown under certain conditions. I found that cold temperatures perturb maternal Wolbachia transmission in host oocytes, which provides a simple explanation for why Wolbachia are less prevalent at temperate latitudes in Drosophila melanogaster fruit fly populations. Genomic analyses and crossing experiments suggest that Wolbachia and hosts from southern Australia have rapidly co-adapted to the temperate climate in only the last ~5,000 years. Wolbachia strains found in other Drosophila species show varying levels of thermal sensitivity, which may help explain why Wolbachia are common in some insect species but not others.

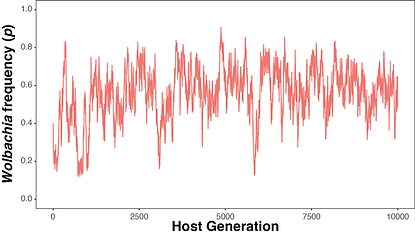
Endosymbiont prevalence fluctuates over time
A small host population can give rise to stochastic fluctuations of endosymbiont frequencies from one host generation to the next. Building on well-established theory, we implemented mathematical models of endosymbiont spread in the publicly available R package symbiontmodeler. Researchers and students can use the package to investigate how factors like host population size contribute to endosymbiont fluctuations over time. Our simulations show that maternal transmission rates play an outsized role in determining the size of stochastic fluctuations in finite host populations.
Endosymbionts alter host fitness
Endosymbionts can have complex effects on the physiology, behavior, and fitness of their hosts. Wolbachia bacteria are found throughout many insect tissues (e.g., the brain), yet we have a poor understanding of how somatic infections affect host fitness. Using a comparative approach, we've found that Wolbachia have pervasive effects on basic aspects of insect physiology and behavior. Thus far, the work has demonstrated that diverse Wolbachia strains diverged put to 50 million years alter the activity levels and thermal preference of their Drosophila hosts.


Evolution at the toxin-binding interface in a predator-prey arms race
Intense coevolution between natural enemies like predators and prey can lead to an arms race. I study adaptation at the molecular interface of toxin-binding in the coevolutionary arms race between garter snakes and their deadly prey, Pacific newts. The work examines how protein evolution and constraints shape escalation of the arms race across western North America.
Convergent evolution of a conserved protein
Arms race coevolution drives rapid adaptation and counter-adaptation, but functional trade-offs may also develop as a consequence. I found that multiple lineages of garter snakes independently evolved resistance to tetrodotoxin (TTX) via a repeated first-step mutation to the Nav1.4 skeletal muscle sodium channel that disrupts toxin-binding, implying that increases in resistance depend on an initial permissive mutation. In highly resistant snakes, accumulating mutations in the channel pore disrupt toxin-binding, but also generate negative trade-offs with Nav1.4 function and muscle performance. These results highlight how costs develop as beneficial mutations accumulate in the arms race. Constraints in Nav1.4 evolution help explain the convergent evolution of TTX resistance in multiple snake populations across western North America.


Asymmetries in the arms race
Reciprocal adaptation is the hallmark of coevolution, but the evolutionary response of each species may not be symmetrical. Levels of snake resistance and newt toxins are closely matched across western North America, implying that phenotypic escalation in both species is a result of the arms race. By the same token, geographic variation in snake TTX resistance shows a clear signature of local adaptation to toxic prey. However, I found that geographic variation in newt toxin levels is best explained by the spatial structure of newt populations, not snake resistance. Newt populations seem to structure variation in prey toxins, which in turn influences local levels of resistance in predators. This unexpected result suggests that neutral processes like gene flow—rather than reciprocal adaptation—represent the greatest source of variation across the geographic mosaic of coevolution.